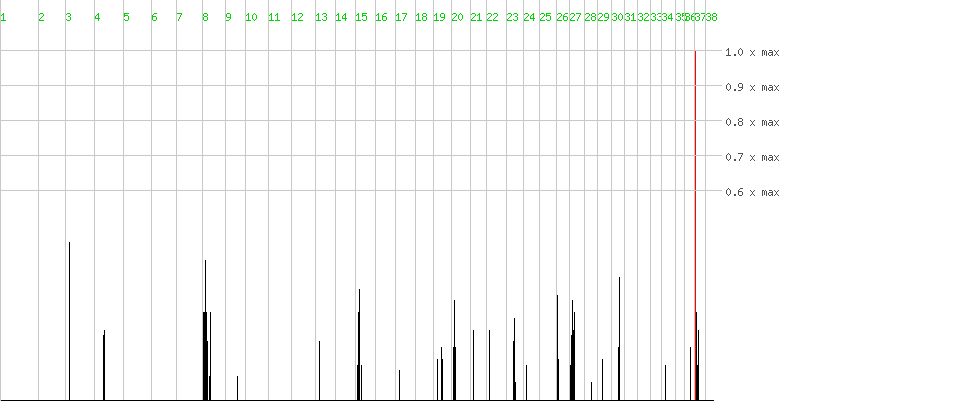
| Click on a chromosome to zoom in. | help tutorial |
| score | chr | from (bp) | to (bp) | from SNP | to SNP | build 37 | |
| broad - use this when you expect some genetic heterogeneity | |||||||
| 60 | 37 | 3902216 | 4045838 | #242511 | #127554 | region | genotypes |
| 60 | 37 | 4245875 | 4508283 | #242532 | #242548 | region | genotypes |
| 48 | 37 | 3721133 | 3902216 | #242499 | #242511 | region | genotypes |
| narrow - use this when all patients are in the same family | |||||||
| 60 | 37 | 3902216 | 4045838 | #242511 | #127554 | region | genotypes |
| 60 | 37 | 4245875 | 4508283 | #242532 | #242548 | region | genotypes |
| 48 | 37 | 3721133 | 3902216 | #242499 | #242511 | region | genotypes |
| analysis settings | ||
| project_name | Canine_hom_map_27_3_12 | change name |
| analysis_name | 300_homo_mapping_ped_2_copy | change name |
| analysis_description | 300 pedigree mapping | |
| max_block_length | 80 | |
| max_score | 60 | |
| user_login | ahramd | |
| homogeneity_required | homogeneity required | |
| lower_limit | 0 | |
| date | 2012-03-30 00:00:00 | |
| exclusion_length | 2000 | |
| cases | 300-007,300-008,300-032 | |
| controls | 300-006 | |