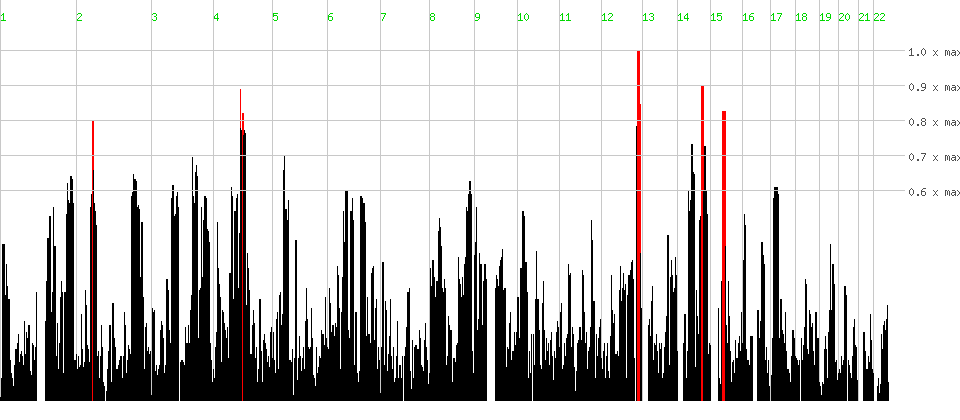
| Click on a chromosome to zoom in. | help tutorial |
| score | chr | from (bp) | to (bp) | from SNP | to SNP | build 37 | |
| broad - use this when you expect some genetic heterogeneity | |||||||
| 160 | 12 | 117080995 | 126921171 | rs953004 | rs722704 | region | genotypes |
| 144 | 14 | 75993940 | 83726679 | rs764187 | rs2124851 | region | genotypes |
| 142 | 4 | 88969925 | 89719368 | rs35491928 | rs1377290 | region | genotypes |
| 135 | 12 | 127672428 | 128075482 | rs950591 | rs2013160 | region | genotypes |
| 132 | 15 | 39227644 | 47875273 | rs1368800 | rs718113 | region | genotypes |
| 131 | 4 | 93000437 | 99033860 | rs1826058 | rs1111541 | region | genotypes |
| 128 | 2 | 52401421 | 54284584 | rs1922192 | rs1363060 | region | genotypes |
| narrow - use this when all patients are in the same family | |||||||
| 160 | 12 | 117080995 | 126921171 | rs953004 | rs722704 | region | genotypes |
| 144 | 14 | 75993940 | 83726679 | rs764187 | rs2124851 | region | genotypes |
| 142 | 4 | 88969925 | 89719368 | rs35491928 | rs1377290 | region | genotypes |
| 135 | 12 | 127672428 | 128075482 | rs950591 | rs2013160 | region | genotypes |
| 132 | 15 | 39227644 | 47875273 | rs1368800 | rs718113 | region | genotypes |
| 131 | 4 | 93000437 | 99033860 | rs1826058 | rs1111541 | region | genotypes |
| 128 | 2 | 52401421 | 54284584 | rs1922192 | rs1363060 | region | genotypes |
| analysis settings | ||
| project_name | Example cutis laxa | change name |
| analysis_name | cutis laxa | change name |
| analysis_description | PubMed | |
| max_block_length | 40 | |
| max_score | 160 | |
| allele_frequencies | controls | |
| user_login | examples | |
| cases | 8106,8107,8108,8109 | |
| controls | 405-5491,407-5803 | |